The fundamental role of nutrition to maintain health, the immune response, and disease prevention, has been known for thousands of years.
The nutritional status of an individual is a key determinant of the susceptibility of the immune system to infection and disease
Emerging insights in the field of nutritional immunology are being investigated with increased urgency.
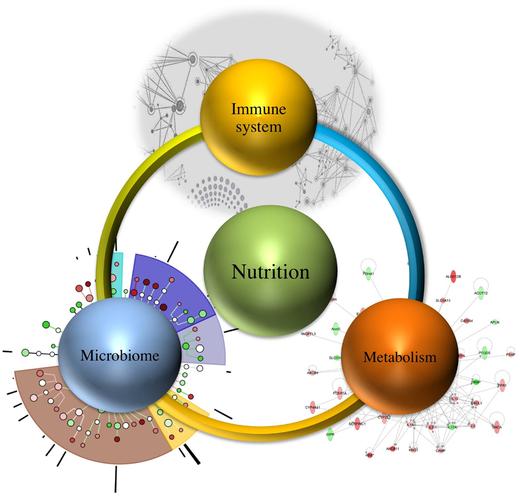
Nutritional immunology is “a massively interacting system of interconnected multistage and multiscale networks that encompass hidden mechanisms by which nutrition, microbiome, metabolism, genetic predisposition, and the immune system interact to delineate health and disease”.
So say researchers at the Nutritional Immunology and Molecular Medicine Laboratory (www.nimml.org) and the Center for Modeling Immunity to Enteric Pathogens, Biocomplexity Institute, Virginia Tech, Blacksburg, VA.
The researchers identified the “potential to discover emerging systems-wide properties at the interface of the immune system, nutrition, microbiome, and metabolism”.
Introduction
2,500 years ago, Hippocrates, famously said “Let food be your medicine and medicine be your food”.
Modern nutritional immunology dates back to the eighteenth century, when the explanation of lymphoid tissue atrophy in malnourished population in England connected nutritional status and immune function.
Epidemiological and clinical data clearly suggest that nutritional deficiencies of essential dietary components, such as vitamins and micronutrients, alter immune competence and increase the risk of infection.
The deficiency of adequate macronutrients and selected micronutrients, such as zinc, selenium, iron, copper, and vitamins A, B-6, C, E, leads to immune deficiency-related infections in children.
Micronutrient deficiencies affect innate immune responses as well as adaptive cellular immune responses.
The immune response is dependent on the nutritional components of food intake, which modulates the induction of regulatory versus effector response at the gut mucosal level.
Recent studies also suggest that the current immune deficiency cases are also the result of increased stress, increased caloric intake, obesity, autoimmunity, allergic disorders, and an aging population.
Unbalanced nutrition, unhealthy lifestyle choices, limited physical activity, and the effect of the environment, in general, compromise the host immune response, increasing the risks of a wide range of diseases.
Recent evidence also suggests the involvement of diet and the role of composition of microbiota in reduced risk of Parkinson’s disease (PD).
The neuroendocrine system can be considered as an important part of the massively interacting multistage networks that define health and wellness.
Whether one is looking at cancer immunology, natural killer cell responses, B cell responses (naïve and memory), T regulatory cell dynamics and T cell responses, research and experience suggests nutrition affects health and disease outcomes.
This post is inspired by the work of many researchers, scientists, and other medical professionals, including those reported in Frontiers In Nutriton.
Footnotes
References
1. Beisel WR. The history of nutritional immunology. J Nutr Immunol (1991) 1(1):5–40.
2. Beisel WR. History of nutritional immunology: introduction and overview1. J Nutr (1992) 122(3S):591.
3. Satyaraj E. Emerging paradigms in immunonutrition. Top Companion Anim Med (2011) 26(1):25–32. doi:10.1053/j.tcam.2011.01.004
PubMed Abstract | CrossRef Full Text | Google Scholar
4. Greicius G, Arulampalam V, Pettersson S. A CLA’s act: feeding away inflammation. Gastroenterology (2004) 127(3):994–6. doi:10.1053/j.gastro.2004.07.038
CrossRef Full Text | Google Scholar
5. Bendich A, Chandra RK. Micronutrients and immune functions. Ann N Y Acad Sci (1990) 587:3–320. doi:10.1111/j.1749-6632.1990.tb00144.x
CrossRef Full Text | Google Scholar
6. Chandra RK. Nutrition and the immune system: an introduction. Am J Clin Nutr (1997) 66(2):460S–3S.
PubMed Abstract | Google Scholar
7. Afacan NJ, Fjell CD, Hancock RE. A systems biology approach to nutritional immunology-focus on innate immunity. Mol Aspects Med (2012) 33(1):14–25. doi:10.1016/j.mam.2011.10.013
CrossRef Full Text | Google Scholar
8. Klingelhoefer L, Reichmann H. Pathogenesis of Parkinson disease – the gut-brain axis and environmental factors. Nat Rev Neurol (2015) 11(11):625–36. doi:10.1038/nrneurol.2015.197
CrossRef Full Text | Google Scholar
9. Kim PS, Levy D, Lee PP. Modeling and simulation of the immune system as a self-regulating network. Methods Enzymol (2009) 467:79–109. doi:10.1016/S0076-6879(09)67004-X
PubMed Abstract | CrossRef Full Text | Google Scholar
10. Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature (2011) 474(7351):327–36. doi:10.1038/nature10213
PubMed Abstract | CrossRef Full Text | Google Scholar
11. Dave M, Higgins PD, Middha S, Rioux KP. The human gut microbiome: current knowledge, challenges, and future directions. Transl Res (2012) 160(4):246–57. doi:10.1016/j.trsl.2012.05.003
PubMed Abstract | CrossRef Full Text | Google Scholar
12. Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A (1998) 95(12):6578–83. doi:10.1073/pnas.95.12.6578
PubMed Abstract | CrossRef Full Text | Google Scholar
13. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature (2007) 449(7164):804–10. doi:10.1038/nature06244
PubMed Abstract | CrossRef Full Text | Google Scholar
14. Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe (2008) 3(4):213–23. doi:10.1016/j.chom.2008.02.015
PubMed Abstract | CrossRef Full Text | Google Scholar
15. Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med (2009) 1(6):ra14–6. doi:10.1126/scitranslmed.3000322
PubMed Abstract | CrossRef Full Text | Google Scholar
16. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature (2006) 444(7122):1027–31. doi:10.1038/nature05414
PubMed Abstract | CrossRef Full Text | Google Scholar
17. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature (2014) 505(7484):559–63. doi:10.1038/nature12820
PubMed Abstract | CrossRef Full Text | Google Scholar
18. Bassaganya-Riera J, Viladomiu M, Pedragosa M, De Simone C, Hontecillas R. Immunoregulatory mechanisms underlying prevention of colitis-associated colorectal cancer by probiotic bacteria. PLoS One (2012) 7(4):e34676. doi:10.1371/journal.pone.0034676
PubMed Abstract | CrossRef Full Text | Google Scholar
19. Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science (2011) 332(6032):970–4. doi:10.1126/science.1198719
PubMed Abstract | CrossRef Full Text | Google Scholar
20. Daniel H, Moghaddas Gholami A, Berry D, Desmarchelier C, Hahne H, Loh G, et al. High-fat diet alters gut microbiota physiology in mice. ISME J (2014) 8(2):295–308. doi:10.1038/ismej.2013.155
PubMed Abstract | CrossRef Full Text | Google Scholar
21. Vermeire S, Noman M, Van Assche G, Baert F, Van Steen K, Esters N, et al. Autoimmunity associated with anti-tumor necrosis factor α treatment in Crohn’s disease: a prospective cohort study. Gastroenterology (2003) 125(1):32–9. doi:10.1016/S0016-5085(03)00701-7
CrossRef Full Text | Google Scholar
22. Bassaganya-Riera J, Viladomiu M, Pedragosa M, De Simone C, Carbo A, Shaykhutdinov R, et al. Probiotic bacteria produce conjugated linoleic acid locally in the gut that targets macrophage PPAR gamma to suppress colitis. PLoS One (2012) 7(2):e31238. doi:10.1371/journal.pone.0031238
CrossRef Full Text | Google Scholar
23. Marion-Letellier R, Déchelotte P, Iacucci M, Ghosh S. Dietary modulation of peroxisome proliferator-activated receptor gamma. Gut (2009) 58(4):586–93. doi:10.1136/gut.2008.162859
PubMed Abstract | CrossRef Full Text | Google Scholar
24. Bassaganya-Riera J, Hontecillas R. Dietary CLA and n-3 PUFA in inflammatory bowel disease. Curr Opin Clin Nutr Metab Care (2010) 13(5):569. doi:10.1097/MCO.0b013e32833b648e
CrossRef Full Text | Google Scholar
25. Borenstein E. Computational systems biology and in silico modeling of the human microbiome. Brief Bioinform (2012) 13(6):769–80. doi:10.1093/bib/bbs022
PubMed Abstract | CrossRef Full Text | Google Scholar
26. Viladomiu M, Hontecillas R, Yuan L, Lu P, Bassaganya-Riera J. Nutritional protective mechanisms against gut inflammation. J Nutr Biochem (2013) 24(6):929–39. doi:10.1016/j.jnutbio.2013.01.006
PubMed Abstract | CrossRef Full Text | Google Scholar
27. Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med (2015) 7(307):ra152–307. doi:10.1126/scitranslmed.aab2271
PubMed Abstract | CrossRef Full Text | Google Scholar
28. Fonseca DM, Hand TW, Han SJ, Gerner MY, Glatman Zaretsky A, Byrd AL, et al. Microbiota-dependent sequelae of acute infection compromise tissue-specific immunity. Cell (2015) 163(2):354–66. doi:10.1016/j.cell.2015.08.030
PubMed Abstract | CrossRef Full Text | Google Scholar
29. Hooper LV. Do symbiotic bacteria subvert host immunity? Nat Rev Microbiol (2009) 7(5):367–74. doi:10.1038/nrmicro2114
PubMed Abstract | CrossRef Full Text | Google Scholar
30. Fagarasan S, Honjo T. Intestinal IgA synthesis: regulation of front-line body defences. Nat Rev Immunol (2003) 3(1):63–72. doi:10.1038/nri982
PubMed Abstract | CrossRef Full Text | Google Scholar
31. Trop TK. Intestinal microbiota, probiotics and prebiotics in inflammatory bowel disease. World J Gastroenterol (2014) 20(33):11505. doi:10.3748/wjg.v20.i33.11505
PubMed Abstract | CrossRef Full Text | Google Scholar
32. Bassaganya-Riera J, DiGuardo M, Viladomiu M, de Horna A, Sanchez S, Einerhand AW, et al. Soluble fibers and resistant starch ameliorate disease activity in interleukin-10-deficient mice with inflammatory bowel disease. J Nutr (2011) 141(7):1318–25. doi:10.3945/jn.111.139022
CrossRef Full Text | Google Scholar
33. Quigley EM. Prebiotics and probiotics their role in the management of gastrointestinal disorders in adults. Nutr Clin Pract (2012) 27(2):195–200. doi:10.1177/0884533611423926
PubMed Abstract | CrossRef Full Text | Google Scholar
34. den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res (2013) 54(9):2325–40. doi:10.1194/jlr.R036012
PubMed Abstract | CrossRef Full Text | Google Scholar
35. Musso G, Gambino R, Cassader M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu Rev Med (2011) 62:361–80. doi:10.1146/annurev-med-012510-175505
PubMed Abstract | CrossRef Full Text | Google Scholar
36. Sakamoto M, Benno Y. Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int J Syst Evol Microbiol (2006) 56(7):1599–605. doi:10.1099/ijs.0.64192-0
PubMed Abstract | CrossRef Full Text | Google Scholar
37. Kverka M, Zakostelska Z, Klimesova K, Sokol D, Hudcovic T, Hrncir T, et al. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin Exp Immunol (2011) 163(2):250–9. doi:10.1111/j.1365-2249.2010.04286.x
PubMed Abstract | CrossRef Full Text | Google Scholar
38. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell (2012) 148(6):1258–70. doi:10.1016/j.cell.2012.01.035
PubMed Abstract | CrossRef Full Text | Google Scholar
39. Johnson R. Immune and endocrine regulation of food intake in sick animals. Domest Anim Endocrinol (1998) 15(5):309–19. doi:10.1016/S0739-7240(98)00031-9
PubMed Abstract | CrossRef Full Text | Google Scholar
40. Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci (2008) 9(1):46–56. doi:10.1038/nrn2297
PubMed Abstract | CrossRef Full Text | Google Scholar
41. Dantzer R. Cytokine-induced sickness behavior: mechanisms and implications. Ann N Y Acad Sci (2001) 933(1):222–34. doi:10.1111/j.1749-6632.2001.tb05827.x
CrossRef Full Text | Google Scholar
42. Palmer CS, Ostrowski M, Balderson B, Christian N, Crowe SM. Glucose metabolism regulates T cell activation, differentiation, and functions. Front Immunol (2015) 6:1. doi:10.3389/fimmu.2015.00001
PubMed Abstract | CrossRef Full Text | Google Scholar
43. Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science (2013) 342(6155):1242454. doi:10.1126/science.1242454
PubMed Abstract | CrossRef Full Text | Google Scholar
44. Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol (2011) 186(6):3299–303. doi:10.4049/jimmunol.1003613
PubMed Abstract | CrossRef Full Text | Google Scholar
45. Guerrant RL, Oriá RB, Moore SR, Oriá MO, Lima AA. Malnutrition as an enteric infectious disease with long-term effects on child development. Nutr Rev (2008) 66(9):487–505. doi:10.1111/j.1753-4887.2008.00082.x
PubMed Abstract | CrossRef Full Text | Google Scholar
46. Philipson CW, Bassaganya-Riera J, Viladomiu M, Pedragosa M, Guerrant RL, Roche JK, et al. The role of peroxisome proliferator-activated receptor gamma in immune responses to enteroaggregative Escherichia coli infection. PLoS One (2013) 8(2):e57812. doi:10.1371/journal.pone.0057812
CrossRef Full Text | Google Scholar
47. Philipson CW, Bassaganya-Riera J, Hontecillas R. Animal models of enteroaggregative Escherichia coli infection. Gut Microbes (2013) 4(4):281–91. doi:10.4161/gmic.24826
CrossRef Full Text | Google Scholar
48. Bolick DT, Kolling GL, Moore JH II, de Oliveira LA, Tung K, Philipson C, et al. Zinc deficiency alters host response and pathogen virulence in a mouse model of enteroaggregative Escherichia coli-induced diarrhea. Gut Microbes (2014) 5(5):618–27. doi:10.4161/19490976.2014.969642
PubMed Abstract | CrossRef Full Text | Google Scholar
49. Wu D, Meydani SN. Age-associated changes in immune and inflammatory responses: impact of vitamin E intervention. J Leukoc Biol (2008) 84(4):900–14. doi:10.1189/jlb.0108023
PubMed Abstract | CrossRef Full Text | Google Scholar
50. Ross AC. Vitamin A and retinoic acid in T cell-related immunity. Am J Clin Nutr (2012) 96(5):1166S–72S. doi:10.3945/ajcn.112.034637
PubMed Abstract | CrossRef Full Text | Google Scholar
51. Haase H, Rink L. Zinc signals and immune function. Biofactors (2014) 40(1):27–40. doi:10.1002/biof.1114
PubMed Abstract | CrossRef Full Text | Google Scholar
52. Fraker PJ, King LE. Reprogramming of the immune system during zinc deficiency. Annu Rev Nutr (2004) 24:277–98. doi:10.1146/annurev.nutr.24.012003.132454
PubMed Abstract | CrossRef Full Text | Google Scholar
53. Calder PC. Very long chain omega-3 (n-3) fatty acids and human health. Eur J Lipid Sci Technol (2014) 116(10):1280–300. doi:10.1002/ejlt.201400025
CrossRef Full Text | Google Scholar
54. Calder PC. Mechanisms of action of (n-3) fatty acids. J Nutr (2012) 142(3):592S–9S. doi:10.3945/jn.111.155259
PubMed Abstract | CrossRef Full Text | Google Scholar
55. Calder PC. Functional roles of fatty acids and their effects on human health. JPEN J Parenter Enteral Nutr (2015) 39(1 Suppl):18S–32S. doi:10.1177/0148607115595980
PubMed Abstract | CrossRef Full Text | Google Scholar
56. Hontecillas R, Wannemeulher MJ, Zimmerman DR, Hutto DL, Wilson JH, Ahn DU, et al. Nutritional regulation of porcine bacterial-induced colitis by conjugated linoleic acid. J Nutr (2002) 132(7):2019–27.
PubMed Abstract | Google Scholar
57. Bassaganya-Riera J, Reynolds K, Martino-Catt S, Cui Y, Hennighausen L, Gonzalez F, et al. Activation of PPAR γ and δ by conjugated linoleic acid mediates protection from experimental inflammatory bowel disease. Gastroenterology (2004) 127(3):777–91. doi:10.1053/j.gastro.2004.06.049
CrossRef Full Text | Google Scholar
58. Bassaganya-Riera J, Hontecillas R. CLA and n-3 PUFA differentially modulate clinical activity and colonic PPAR-responsive gene expression in a pig model of experimental IBD. Clin Nutr (2006) 25(3):454–65. doi:10.1016/j.clnu.2005.12.008
PubMed Abstract | CrossRef Full Text | Google Scholar
59. Evans NP, Misyak SA, Schmelz EM, Guri AJ, Hontecillas R, Bassaganya-Riera J. Conjugated linoleic acid ameliorates inflammation-induced colorectal cancer in mice through activation of PPARγ. J Nutr (2010) 140(3):515–21. doi:10.3945/jn.109.115642
CrossRef Full Text | Google Scholar
60. Bassaganya-Riera J, Pogranichniy RM, Jobgen SC, Halbur PG, Yoon KJ, O’Shea M, et al. Conjugated linoleic acid ameliorates viral infectivity in a pig model of virally induced immunosuppression. J Nutr (2003) 133(10):3204–14.
PubMed Abstract | Google Scholar
61. Bassaganya-Riera J, Hontecillas R, Zimmerman DR, Wannemuehler MJ. Long-term influence of lipid nutrition on the induction of CD8+ responses to viral or bacterial antigens. Vaccine (2002) 20(9):1435–44. doi:10.1016/S0264-410X(01)00465-0
CrossRef Full Text | Google Scholar
62. Bassaganya-Riera J, Hontecillas R, Horne WT, Sandridge M, Herfarth HH, Bloomfeld R, et al. Conjugated linoleic acid modulates immune responses in patients with mild to moderately active Crohn’s disease. Clin Nutr (2012) 31(5):721–7. doi:10.1016/j.clnu.2012.03.002
PubMed Abstract | CrossRef Full Text | Google Scholar
63. Abedi V, Lu P, Hontecillas R, Verma M, Vess G, Philipson C, et al. Phase III placebo-controlled, randomized clinical trial with synthetic Crohn’s disease patients to evaluate treatment response. Emerging Trends in Computational Biology, Bioinformatics, and Systems Biology – Systems & Applications. Elsevier/MK (2016) (in press).
64. Guri AJ, Hontecillas R, Si H, Liu D, Bassaganya-Riera J. Dietary abscisic acid ameliorates glucose tolerance and obesity-related inflammation in db/db mice fed high-fat diets. Clin Nutr (2007) 26(1):107–16. doi:10.1016/j.clnu.2006.07.008
PubMed Abstract | CrossRef Full Text | Google Scholar
65. Guri AJ, Hontecillas R, Bassaganya-Riera J. Abscisic acid ameliorates experimental IBD by downregulating cellular adhesion molecule expression and suppressing immune cell infiltration. Clin Nutr (2010) 29(6):824–31. doi:10.1016/j.clnu.2010.02.009
PubMed Abstract | CrossRef Full Text | Google Scholar
66. Guri AJ, Misyak SA, Hontecillas R, Hasty A, Liu D, Si H, et al. Abscisic acid ameliorates atherosclerosis by suppressing macrophage and CD4+ T cell recruitment into the aortic wall. J Nutr Biochem (2010) 21(12):1178–85. doi:10.1016/j.jnutbio.2009.10.003
PubMed Abstract | CrossRef Full Text | Google Scholar
67. Bassaganya-Riera J, Guri AJ, Lu P, Climent M, Carbo A, Sobral BW, et al. Abscisic acid regulates inflammation via ligand-binding domain-independent activation of peroxisome proliferator-activated receptor γ. J Biol Chem (2011) 286(4):2504–16. doi:10.1074/jbc.M110.160077
CrossRef Full Text | Google Scholar
68. Hontecillas R, Roberts PC, Carbo A, Vives C, Horne WT, Genis S, et al. Dietary abscisic acid ameliorates influenza-virus-associated disease and pulmonary immunopathology through a PPARγ-dependent mechanism. J Nutr Biochem (2013) 24(6):1019–27. doi:10.1016/j.jnutbio.2012.07.010
PubMed Abstract | CrossRef Full Text | Google Scholar
69. Magnone M, Ameri P, Salis A, Andraghetti G, Emionite L, Murialdo G, et al. Microgram amounts of abscisic acid in fruit extracts improve glucose tolerance and reduce insulinemia in rats and in humans. FASEB J (2015) 29(12):4783–93. doi:10.1096/fj.15-277731
PubMed Abstract | CrossRef Full Text | Google Scholar
70. Kubena KS, McMurray DN. Nutrition and the immune system: a review of nutrient-nutrient interactions. J Am Diet Assoc (1996) 96(11):1156–64. doi:10.1016/S0002-8223(96)00297-0
CrossRef Full Text | Google Scholar
71. Lacroix S, Lauria M, Scott-Boyer MP, Marchetti L, Priami C, Caberlotto L. Systems biology approaches to study the molecular effects of caloric restriction and polyphenols on aging processes. Genes Nutr (2015) 10(6):58. doi:10.1007/s12263-015-0508-9
PubMed Abstract | CrossRef Full Text | Google Scholar
72. Gardy JL, Lynn DJ, Brinkman FS, Hancock RE. Enabling a systems biology approach to immunology: focus on innate immunity. Trends Immunol (2009) 30(6):249–62. doi:10.1016/j.it.2009.03.009
PubMed Abstract | CrossRef Full Text | Google Scholar
73. Kaput J, Rodriguez RL. Nutritional genomics: the next frontier in the postgenomic era. Physiol Genomics (2004) 16(2):166–77. doi:10.1152/physiolgenomics.00107.2003
PubMed Abstract | CrossRef Full Text | Google Scholar
74. Allison DB, Bassaganya-Riera J, Burlingame B, Brown AW, le Coutre J, Dickson SL, et al. Goals in nutrition Science 2015-2020. Front Nutr (2015) 2:26. doi:10.3389/fnut.2015.00026
CrossRef Full Text | Google Scholar
75. Brennan L. Metabolomics in nutrition research: current status and perspectives. Biochem Soc Trans (2013) 41(2):670–3. doi:10.1042/BST20120350
PubMed Abstract | CrossRef Full Text | Google Scholar
76. Bakker GC, van Erk MJ, Pellis L, Wopereis S, Rubingh CM, Cnubben NH, et al. An antiinflammatory dietary mix modulates inflammation and oxidative and metabolic stress in overweight men: a nutrigenomics approach. Am J Clin Nutr (2010) 91(4):1044–59. doi:10.3945/ajcn.2009.28822
PubMed Abstract | CrossRef Full Text | Google Scholar
77. Carbo A, Hontecillas R, Andrew T, Eden K, Mei Y, Hoops S, et al. Computational modeling of heterogeneity and function of CD4+ T cells. Front Cell Dev Biol (2014) 2:31. doi:10.3389/fcell.2014.00031
PubMed Abstract | CrossRef Full Text | Google Scholar
78. Mei Y, Abedi V, Carbo A, Zhang X, Lu P, Philipson C, et al. Multiscale modeling of mucosal immune responses. BMC Bioinformatics (2015) 16(Suppl 12):S2. doi:10.1186/1471-2105-16-S12-S2
PubMed Abstract | CrossRef Full Text | Google Scholar
79. Wendelsdorf K, Bassaganya-Riera J, Hontecillas R, Eubank S. Model of colonic inflammation: immune modulatory mechanisms in inflammatory bowel disease. J Theor Biol (2010) 264(4):1225–39. doi:10.1016/j.jtbi.2010.03.027
PubMed Abstract | CrossRef Full Text | Google Scholar
80. Ghosh S, Matsuoka Y, Asai Y, Hsin KY, Kitano H. Software for systems biology: from tools to integrated platforms. Nat Rev Genet (2011) 12(12):821–32. doi:10.1038/nrg3096
PubMed Abstract | CrossRef Full Text | Google Scholar
81. Masoudi-Nejad A, Bidkhori G, Hosseini Ashtiani S, Najafi A, Bozorgmehr JH, Wang E. Cancer systems biology and modeling: microscopic scale and multiscale approaches. Semin Cancer Biol (2015) 30C:60–9. doi:10.1016/j.semcancer.2014.03.003
PubMed Abstract | CrossRef Full Text | Google Scholar
82. Castiglione F, Pappalardo F, Bianca C, Russo G, Motta S. Modeling biology spanning different scales: an open challenge. Biomed Res Int (2014) 2014:902545. doi:10.1155/2014/902545
PubMed Abstract | CrossRef Full Text | Google Scholar
83. Dwivedi G, Fitz L, Hegen M, Martin SW, Harrold J, Heatherington A, et al. A multiscale model of interleukin-6-mediated immune regulation in Crohn’s disease and its application in drug discovery and development. CPT Pharmacometrics Syst Pharmacol (2014) 3:e89. doi:10.1038/psp.2013.64
PubMed Abstract | CrossRef Full Text | Google Scholar
84. Mei Y, Carbo A, Hontecillas R, Hoops S, Liles N, Pinyi L, et al. ENISI MSM: a novel multi-scale modeling platform for computational immunology. 2014 IEEE International Conference on Bioinformatics and Biomedicine. Belfast: IEEE (2014). p. 391–6.
85. Sütterlin T, Kolb C, Dickhaus H, Jäger D, Grabe N. Bridging the scales: semantic integration of quantitative SBML in graphical multi-cellular models and simulations with EPISIM and COPASI. Bioinformatics (2013) 29(2):223–9. doi:10.1093/bioinformatics/bts659
PubMed Abstract | CrossRef Full Text | Google Scholar
86. Mc Auley MT, Proctor CJ, Corfe BM, Cuskelly GCJ, Mooney KM. Nutrition research and the impact of computational systems biology. J Comput Sci Syst Biol (2013) 6(5):271–85. doi:10.4172/jcsb.1000122
CrossRef Full Text | Google Scholar
87. Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics (2003) 19(4):524–31. doi:10.1093/bioinformatics/btg015
PubMed Abstract | CrossRef Full Text | Google Scholar
88. Ivanciuc O, Gendel SM, Power TD, Schein CH, Braun W. AllerML: markup language for allergens. Regul Toxicol Pharmacol (2011) 60(1):151–60. doi:10.1016/j.yrtph.2011.03.006
PubMed Abstract | CrossRef Full Text | Google Scholar
89. Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform (2008):67–79.
PubMed Abstract | Google Scholar
90. Information NCFB. Medical Subject Headings. Available from: http://www.ncbi.nlm.nih.gov/mesh
91. Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res (2004) 32:D267–70. doi:10.1093/nar/gkh061
PubMed Abstract | CrossRef Full Text | Google Scholar
92. Zhu F, Shi Z, Qin C, Tao L, Liu X, Xu F, et al. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res (2012) 40(Database issue):D1128–36. doi:10.1093/nar/gkr797
PubMed Abstract | CrossRef Full Text | Google Scholar
93. Zhu X, Kruhlak NL. Construction and analysis of a human hepatotoxicity database suitable for QSAR modeling using post-market safety data. Toxicology (2014) 321:62–72. doi:10.1016/j.tox.2014.03.009
PubMed Abstract | CrossRef Full Text | Google Scholar
94. Liu Y, Hu B, Fu C, Chen X. DCDB: drug combination database. Bioinformatics (2010) 26(4):587–8. doi:10.1093/bioinformatics/btp697
PubMed Abstract | CrossRef Full Text | Google Scholar
95. Arvidson KB. FDA toxicity databases and real-time data entry. Toxicol Appl Pharmacol (2008) 233(1):17–9. doi:10.1016/j.taap.2007.12.033
PubMed Abstract | CrossRef Full Text | Google Scholar
96. Kelder T, Summer G, Caspers M, van Schothorst EM, Keijer J, Duivenvoorde L, et al. White adipose tissue reference network: a knowledge resource for exploring health-relevant relations. Genes Nutr (2015) 10:439. doi:10.1007/s12263-014-0439-x
PubMed Abstract | CrossRef Full Text | Google Scholar
97. Ramsundar B, Steven K, Patrick R, Dale W, David K, Vijay P. Massively multitask networks for drug discovery (2015). arXiv preprint arXiv:1502.02072.
98. Lu P, Abedi V, Mei Y, Hontecillas R, Hoops S, Carbo A, et al. Supervised learning methods in modeling of CD4+ T cell heterogeneity. BioData Min (2015) 8:27. doi:10.1186/s13040-015-0060-6
PubMed Abstract | CrossRef Full Text | Google Scholar
99. Lu P, Abedi V, Mei Y, Hontecillas R, Philipson C, Hoops S, et al. Supervised learning with artificial neural network in modeling of cell differentiation process. In: Tran QN, Arabnia H, editors. Emerging Trends in Computational Biology, Bioinformatics, and Systems Biology. Burlington, MA: Elsevier (2015). 674 p.
100. Abedi V, Hoops S, Hontecillas R, Carbo A, Philipson C, Viladomiu M, et al. Computational Immunology: Models and Tools. In: Bassaganya-Riera J, editor. Boston, MA: Elsevier (2015). 210 p.
101. Philipson CW, Bassaganya-Riera J, Viladomiu M, Kronsteiner B, Abedi V, Hoops S, et al. Modeling the regulatory mechanisms by which NLRX1 modulates innate immune responses to Helicobacter pylori infection. PLoS One (2015) 10(9):e0137839. doi:10.1371/journal.pone.0137839
PubMed Abstract | CrossRef Full Text | Google Scholar
102. Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature (2013) 496(7446):461–8. doi:10.1038/nature11981
PubMed Abstract | CrossRef Full Text | Google Scholar
103. An G, Mi Q, Dutta-Moscato J, Vodovotz Y. Agent-based models in translational systems biology. Wiley Interdiscip Rev Syst Biol Med (2009) 1(2):159–71. doi:10.1002/wsbm.45
CrossRef Full Text | Google Scholar
104. Mei Y, Hontecillas R, Xiaoying Z, Bisset K, Eubank S, Hoops S, et al. ENISI visual, an agent-based simulator for modeling gut immunity. Bioinformatics and Biomedicine (BIBM), 2012 IEEE International Conference on. Philadelphia, PA: IEEE (2012).
105. Alam M, Abedi V, Bassaganya-Riera J, Wendelsdorf K, Bisset K, Deng X, et al. Computational Immunology: Models and Tools. Boston, MA: Elsevier (2015).
106. Vida Abedi RH, Stefan H, Nathan L, Adria C, Pinyi L, Casandra P, et al. ENISI multiscale modeling of mucosal immune responses driven by high performance computing. IEEE International Conference on Bioinformatics and Biomedicine (BIBM 2015). Washington, DC (2015).
107. Bisset K, Alam MM, Bassaganya-Riera J, Carbo A, Eubank S, Hontecillas R, et al. High-performance interaction-based simulation of gut immunopathologies with enteric immunity simulator (ENISI). Parallel & Distributed Processing Symposium (IPDPS), 2012 IEEE 26th International. Shanghai: IEEE (2012).
108. Mei Y, Carbo A, Hontecillas R, Bassaganya-Riera J. ENISI SDE: a novel web-based stochastic modeling tool for computational biology. Bioinformatics and Biomedicine (BIBM), 2013 IEEE International Conference on. Shanghai: IEEE (2013).
109. Wendeldorf KV, Bassaganya-Riera J, Bisset K, Eubank S, Hontecillas R, Marathe M. Enteric immunity simulator: a tool for in silico study of gut immunopathologies. Bioinformatics and Biomedicine (BIBM), 2011 IEEE International Conference on. Atlanta, GA: IEEE (2011).
110. Wendelsdorf KV, Alam M, Bassaganya-Riera J, Bisset K, Eubank S, Hontecillas R, et al. ENteric Immunity SImulator: a tool for in silico study of gastroenteric infections. IEEE Trans Nanobioscience (2012) 11(3):273–88. doi:10.1109/TNB.2012.2211891
PubMed Abstract | CrossRef Full Text | Google Scholar
111. Ekins S, Mestres J, Testa B. In silico pharmacology for drug discovery: methods for virtual ligand screening and profiling. Br J Pharmacol (2007) 152(1):9–20. doi:10.1038/sj.bjp.0707305
PubMed Abstract | CrossRef Full Text | Google Scholar
112. Romero K, Ito K, Rogers JA, Polhamus D, Qiu R, Stephenson D, et al. The future is now: model-based clinical trial design for Alzheimer’s disease. Clin Pharmacol Ther (2015) 97(3):210–4. doi:10.1002/cpt.16
CrossRef Full Text | Google Scholar
113. Brown D, Namas RA, Almahmoud K, Zaaqoq A, Sarkar J, Barclay DA, et al. Trauma in silico: individual-specific mathematical models and virtual clinical populations. Sci Transl Med (2015) 7(285):ra61–285. doi:10.1126/scitranslmed.aaa3636
PubMed Abstract | CrossRef Full Text | Google Scholar
114. Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature (2012) 488(7410):178–84. doi:10.1038/nature11319
PubMed Abstract | CrossRef Full Text | Google Scholar
115. Lu P, Bevan DR, Lewis SN, Hontecillas R, Bassaganya-Riera J. Molecular modeling of lanthionine synthetase component C-like protein 2: a potential target for the discovery of novel type 2 diabetes prophylactics and therapeutics. J Mol Model (2011) 17(3):543–53. doi:10.1007/s00894-010-0748-y
PubMed Abstract | CrossRef Full Text | Google Scholar
116. Carbo A, Hontecillas R, Cooper J, Gandour RD, Ehrich M, Bassaganya-Riera J. Mo1691 lanthionine synthetase C-like receptor 2 (LANCL2): a novel therapeutic target for inflammatory bowel disease. Gastroenterology (2015) 148(4):S–686. doi:10.1016/S0016-5085(15)32321-0
CrossRef Full Text | Google Scholar
117. Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Org E, Parks B, et al. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat Commun (2014) 5:4500. doi:10.1038/ncomms5500
PubMed Abstract | CrossRef Full Text | Google Scholar
118. Marx V. Biology: the big challenges of big data. Nature (2013) 498(7453):255–60. doi:10.1038/498255a
CrossRef Full Text | Google Scholar
119. Fan J, Han F, Liu H. Challenges of big data analysis. Natl Sci Rev (2014) 1(2):293–314. doi:10.1093/nsr/nwt032
PubMed Abstract | CrossRef Full Text | Google Scholar
120. Trelles O, Prins P, Snir M, Jansen RC. Big data, but are we ready? Nat Rev Genet (2011) 12(3):224–224. doi:10.1038/nrg2857-c1
CrossRef Full Text | Google Scholar
121. Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, et al. COPASI – a complex pathway simulator. Bioinformatics (2006) 22(24):3067–74. doi:10.1093/bioinformatics/btl485
CrossRef Full Text | Google Scholar
122. Mendes P, Stefan H, Sven S, Ralph G, Joseph D, Ursula K. Computational modeling of biochemical networks using COPASI. Systems Biology (2009):17–59.
123. Mei Y, Carbo A, Hoops S, Hontecillas R, Bassaganya-Riera J. ENISI SDE: a web-based tool for modeling stochastic processes. IEEE/ACM Trans Comput Biol Bioinform (2015) 12(2):289–97. doi:10.1109/TCBB.2014.2351823
CrossRef Full Text | Google Scholar
124. Carbo A, Hontecillas R, Kronsteiner B, Viladomiu M, Pedragosa M, Lu P, et al. Systems modeling of molecular mechanisms controlling cytokine-driven CD4+ T cell differentiation and phenotype plasticity. PLoS Comput Biol (2013) 9(4):e1003027. doi:10.1371/journal.pcbi.1003027
PubMed Abstract | CrossRef Full Text | Google Scholar
125. Carbo A, Olivares-Villagómez D, Hontecillas R, Bassaganya-Riera J, Chaturvedi R, Piazuelo MB, et al. Systems modeling of the role of IL-21 in the maintenance of effector CD4+ T cell responses during Helicobacter pylori infection. MBio (2014) 5(4):e1243–1214. doi:10.1128/mBio.01243-14
CrossRef Full Text | Google Scholar
126. Carbo A, Bassaganya-Riera J, Pedragosa M, Viladomiu M, Marathe M, Eubank S, et al. Predictive computational modeling of the mucosal immune responses during Helicobacter pylori infection. PLoS One (2013) 8(9):e73365. doi:10.1371/journal.pone.0073365
PubMed Abstract | CrossRef Full Text | Google Scholar
127. Kronsteiner B, Bassaganya-Riera J, Philipson C, Viladomiu M, Carbo A, Pedragosa M, et al. Helicobacter pylori infection in a pig model is dominated by Th1 and cytotoxic CD8+ T cell responses. Infect Immun (2013) 81(10):3803–13. doi:10.1128/IAI.00660-13
PubMed Abstract | CrossRef Full Text | Google Scholar
128. Kronsteiner B, Bassaganya-Riera J, Philipson N, Hontecillas R. Novel insights on the role of CD8+ T cells and cytotoxic responses during Helicobacter pylori infection. Gut Microbes (2014) 5(3):357–62. doi:10.4161/gmic.28899
PubMed Abstract | CrossRef Full Text | Google Scholar
129. Philipson CW, Bassaganya-Riera J, Hontecillas R. Animal models of enteroaggregative Escherichia coli infection. Gut Microbes (2013) 4(4):281–91. doi:10.4161/gmic.24826
PubMed Abstract | CrossRef Full Text | Google Scholar
130. Bolick DT, Roche JK, Hontecillas R, Bassaganya-Riera J, Nataro JP, Guerrant RL. Enteroaggregative Escherichia coli strain in a novel weaned mouse model: exacerbation by malnutrition, biofilm as a virulence factor and treatment by nitazoxanide. J Med Microbiol (2013) 62(Pt 6):896–905. doi:10.1099/jmm.0.046300-0
PubMed Abstract | CrossRef Full Text | Google Scholar
131. Bassaganya-Riera J, Hontecillas R, Abedi V, Carbo A, Philipson C, Hoops S. Computational Immunology: Models and Tools. Boston, MA: Elsevier (2015).
132. Gorenshteyn D, Zaslavsky E, Fribourg M, Park CY, Wong AK, Tadych A, et al. Interactive big data resource to elucidate human immune pathways and diseases. Immunity (2015) 43(3):605–14. doi:10.1016/j.immuni.2015.08.014
PubMed Abstract | CrossRef Full Text | Google Scholar
Keywords: nutritional immunology, nutrition, systems biology, informatics, computational modeling, big data, complex systems
Citation: Verma M, Hontecillas R, Abedi V, Leber A, Tubau-Juni N, Philipson C, Carbo A and Bassaganya-Riera J (2016) Modeling-Enabled Systems Nutritional Immunology. Front. Nutr. 3:5. doi: 10.3389/fnut.2016.00005
This information is shared under Creative Commons Attribution License (CC BY).